 A male and female Amboseli baboon maintaining close spatial proximity, an affiliative behavior important to establishing, maintaining, and strengthening social bonds in non-human primates.
A male and female Amboseli baboon maintaining close spatial proximity, an affiliative behavior important to establishing, maintaining, and strengthening social bonds in non-human primates.
Genetic ancestry effects on affiliative male-female social behavior in baboons
Social relationships, both within and between sexes, are important predictors of fitness in both humans and other social mammals. In monogamous species, long-term social bonds between sexual partners have been shown to predict shorter interbirth intervals, increased offspring number, and improved offspring survival. Similarly, in some group-living primates, females also compete for access to males outside of mating contexts, suggesting that social bonds with males are themselves an important social resource. Understanding why some individuals successfully form opposite-sex social bonds, while others do not, is therefore an important part of understanding the fitness benefits of these relationships.
In contrast to male-female bond formation in monogamous species, little is known about genetic contributions to these relationships in group-living species. This project investigates the association between genetic ancestry and male-female social bonds, including the additive effects of male and female individual genetic ancestry and the role of characteristics defined by the pair, including ancestry-related assortativity. We analyze these effects in the context of social and demographic sources of variance. Check out our Animal Behaviour paper to learn what we found!
Social relationships, both within and between sexes, are important predictors of fitness in both humans and other social mammals. In monogamous species, long-term social bonds between sexual partners have been shown to predict shorter interbirth intervals, increased offspring number, and improved offspring survival. Similarly, in some group-living primates, females also compete for access to males outside of mating contexts, suggesting that social bonds with males are themselves an important social resource. Understanding why some individuals successfully form opposite-sex social bonds, while others do not, is therefore an important part of understanding the fitness benefits of these relationships.
In contrast to male-female bond formation in monogamous species, little is known about genetic contributions to these relationships in group-living species. This project investigates the association between genetic ancestry and male-female social bonds, including the additive effects of male and female individual genetic ancestry and the role of characteristics defined by the pair, including ancestry-related assortativity. We analyze these effects in the context of social and demographic sources of variance. Check out our Animal Behaviour paper to learn what we found!
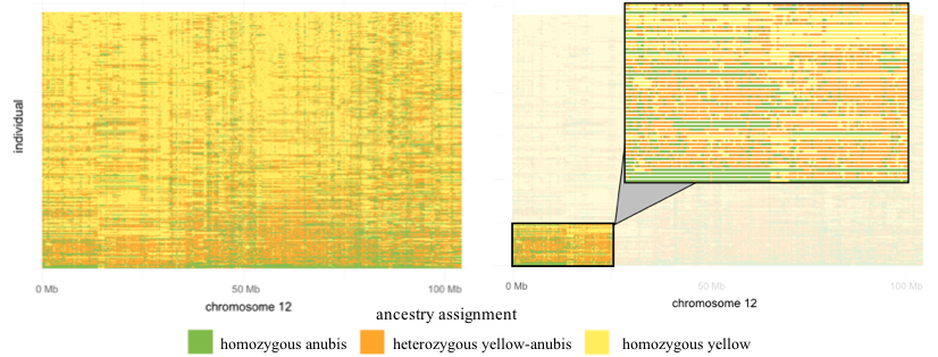 POPULATION-LEVEL LOCAL ANCESTRY. Local ancestry estimates for 290 baboons in a 100 Mb chunk of chromosome 12. Each horizontal bar corresponds to one individual, organized from top to bottom from the most yellow to the most anubis. Figure on the right highlights the first 25 Mb of chromosome 12 for the 50 baboons with the most anubis ancestry on chromosome 12.
POPULATION-LEVEL LOCAL ANCESTRY. Local ancestry estimates for 290 baboons in a 100 Mb chunk of chromosome 12. Each horizontal bar corresponds to one individual, organized from top to bottom from the most yellow to the most anubis. Figure on the right highlights the first 25 Mb of chromosome 12 for the 50 baboons with the most anubis ancestry on chromosome 12.
Natural hybridization has been documented across a wide range of plant and animal taxa. However, many taxa that hybridize maintain clearly differentiated genomes and/or phenotypes despite our expectation that hybridization should have a homogenizing effect and ultimately lead to the erosion of species’ differences. These observations present an intriguing puzzle: what forces maintain phenotypic and genetic integrity in the face of hybridization?
Previous research on the Amboseli baboon hybrids has identified no clear barriers to admixture. Indeed, previous research suggests several advantages associated with anubis ancestry (Charpentier et al. 2008, Tung et al. 2012, Fogel et al. 2021 above). However, both the narrowness of the hybrid zone relative to the parental species’ ranges (Charpentier et al. 2012) and genomic evidence for a long history of intermittent admixture (Wall et al. 2016, Rogers et al. 2019) suggest that gene flow is restricted.
Genomic evidence for selection against introgression
In collaboration with Tauras Vilaglys (previous Tung Lab PhD student), this project investigates evidence for selection against admixture in the Amboseli population using genomic data, tests whether signatures of selection against admixture in archaic hominins generalize to other primates, and tests whether selection is stronger in regions of the genome that exhibit regulatory divergence. Check out our Science paper to learn what we found!
Genetic incompatibilities and assortative mating
To identify specific candidate genetic loci involved in restricting gene flow, this project tests for ancestry-associated interchromosomal linkage disequilibrium (LD) patterns. Such LD patterns could arise due to: (1) ancestry-based assortative mating (preferential mating between phenotypically similar individuals, when those phenotypes have a genetic basis and differ by ancestry); (2) intrinsic, postzygotic Bateson-Dobzhansky-Muller incompatibilities (BDMIs: alleles at two or more loci that evolved in separate parental populations, but negatively interact when in the same genome); or (3) extrinsic, postzygotic selection against hybrid phenotypes. To investigate these alternatives, I am linking the genetic data to long-term data on mating patterns to ask whether assortative mating could explain interchromosomal LD pairs, or whether they are best accounted for by an alternative explanation. Stay tuned to see what we found!
Previous research on the Amboseli baboon hybrids has identified no clear barriers to admixture. Indeed, previous research suggests several advantages associated with anubis ancestry (Charpentier et al. 2008, Tung et al. 2012, Fogel et al. 2021 above). However, both the narrowness of the hybrid zone relative to the parental species’ ranges (Charpentier et al. 2012) and genomic evidence for a long history of intermittent admixture (Wall et al. 2016, Rogers et al. 2019) suggest that gene flow is restricted.
Genomic evidence for selection against introgression
In collaboration with Tauras Vilaglys (previous Tung Lab PhD student), this project investigates evidence for selection against admixture in the Amboseli population using genomic data, tests whether signatures of selection against admixture in archaic hominins generalize to other primates, and tests whether selection is stronger in regions of the genome that exhibit regulatory divergence. Check out our Science paper to learn what we found!
Genetic incompatibilities and assortative mating
To identify specific candidate genetic loci involved in restricting gene flow, this project tests for ancestry-associated interchromosomal linkage disequilibrium (LD) patterns. Such LD patterns could arise due to: (1) ancestry-based assortative mating (preferential mating between phenotypically similar individuals, when those phenotypes have a genetic basis and differ by ancestry); (2) intrinsic, postzygotic Bateson-Dobzhansky-Muller incompatibilities (BDMIs: alleles at two or more loci that evolved in separate parental populations, but negatively interact when in the same genome); or (3) extrinsic, postzygotic selection against hybrid phenotypes. To investigate these alternatives, I am linking the genetic data to long-term data on mating patterns to ask whether assortative mating could explain interchromosomal LD pairs, or whether they are best accounted for by an alternative explanation. Stay tuned to see what we found!
Adaptive introgression and its phenotypic consequences in hybridizing baboons
Hybridization between divergent taxa has classically been considered “a reproductive mistake” that results in less fit offspring (Mallet 2005). Nevertheless, introgression may also confer adaptive benefits. For instance, evidence from our own lineage suggests that while most archaically introduced loci were likely deleterious or neutral, some variants may have helped humans adapt to new ecological pressures as they expanded across the globe. However, we do not yet understand whether the relative costs and benefits identified in studies of hominin admixture are typical of primates more broadly.
In this project, I am investigating signatures of ancestry-related adaptation and phenotypic diversity in the Amboseli hybrids as well as 14 other populations spanning the yellow-anubis contact zone in southern Kenya (Charpentier et al. 2012). I will test the hypotheses that (i) hybridization facilitates the transfer of a small number of advantageous genetic variants between yellow baboons and anubis baboons and (ii) genetic ancestry predicts hair color and maturation timing, traits that may be important in mate competition, fertility, or survival. This work is supported by a Doctoral Dissertation Research Improvement Grant from the National Science Foundation (thank you NSF!).
Hybridization between divergent taxa has classically been considered “a reproductive mistake” that results in less fit offspring (Mallet 2005). Nevertheless, introgression may also confer adaptive benefits. For instance, evidence from our own lineage suggests that while most archaically introduced loci were likely deleterious or neutral, some variants may have helped humans adapt to new ecological pressures as they expanded across the globe. However, we do not yet understand whether the relative costs and benefits identified in studies of hominin admixture are typical of primates more broadly.
In this project, I am investigating signatures of ancestry-related adaptation and phenotypic diversity in the Amboseli hybrids as well as 14 other populations spanning the yellow-anubis contact zone in southern Kenya (Charpentier et al. 2012). I will test the hypotheses that (i) hybridization facilitates the transfer of a small number of advantageous genetic variants between yellow baboons and anubis baboons and (ii) genetic ancestry predicts hair color and maturation timing, traits that may be important in mate competition, fertility, or survival. This work is supported by a Doctoral Dissertation Research Improvement Grant from the National Science Foundation (thank you NSF!).
